Clinical microbiology laboratories are vital to the prevention, diagnosis, and treatment of infectious diseases caused by microorganisms (e.g., bacteria, fungi).
Image Credit: Pakpoom Nunjui/Shuttterstock.com
Microbiologists frequently cultivate clinical samples on solid media in Petri dishes to determine which bacteria cause an infection, after which they select and develop bacterial colonies according to the chosen identification procedure.
Since it is more accurate, quicker, and less labor-intensive than previous approaches, mass spectrometry (MS) has become the most widely used methodology for detecting bacteria and fungus in the last two decades.
Historically, colony selection and depositing were done by hand with loops, toothpicks, or pipette tips. However, skilled technicians are usually necessary to select the correct colony.
Collaborative robot advancements have opened up new possibilities for laboratory automation. Collaborative robots are built for safe robot applications near the operator. Their built-in force sensors detect potential collisions and allow the user to manipulate the robot by gripping and moving individual pieces.
As a result, operators can demonstrate a new task rather than programming the robot. This method is known as kinesthetic teaching and is a type of learning-from-demonstration (LfD) method.
The two proven ways for demonstrating fine movements are teleoperation and cooperative robot tools (CRT). However, to manage the robot, they require extra input devices, which novice operators are unable to use effectively.
Furthermore, it has been demonstrated that, while there are two ways perform better, kinesthetic teaching can also attain sub-millimeter accuracy.
As a result, by using kinesthetic teaching to teach diverse activities (such as colony picking), it may be possible to effectively incorporate collaborative robots into the laboratory setting.
In work published in the MDPI journal sensors, researchers looked at whether collaborative robots could be employed in the lab to perform small tasks that were demonstrated by the operator using kinesthetic teaching.
Methodology
The experimental procedure included a Panda robot manipulator, a MALDI Biotyper mass spectrometer, a SteriMax smart thermal sterilizer, and disposable MALDI target plates MBT Biotarget 96.
An RGB camera a2A3840-45ucPRO, a 2D laser profile measurement system ZG2-WDS8T, a force sensor Nano17, a feedback 360° high-speed servo motor, and a stainless steel needle made up the end-effector tool coupled to the robot manipulator (Figure 1).
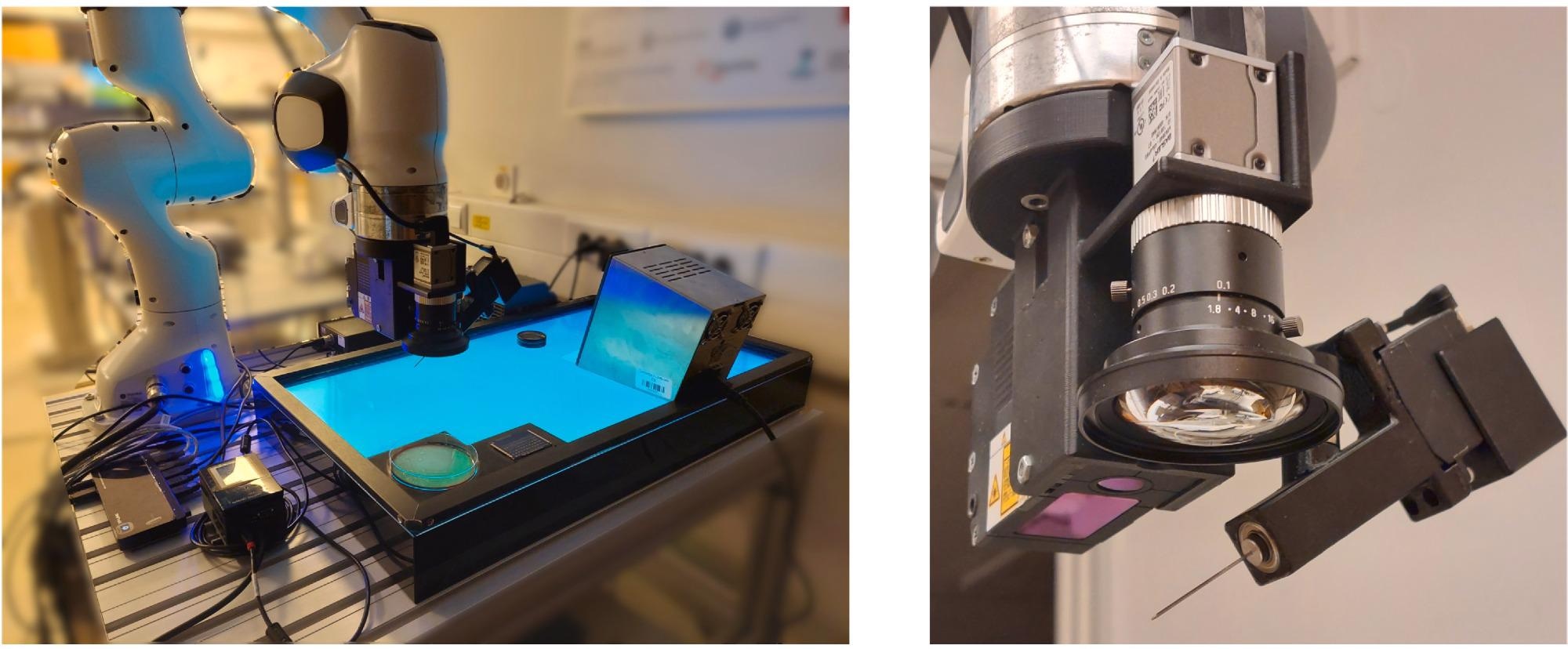
Figure 1. Overview of the experiment setup (left) and the end-effector tool (right). Image Credit: Baumkircher, et al., 2022
The RGB camera took a picture of the Petri dish (Figure 2a), which allowed the operator to estimate the location of the chosen bacterial colony. The area surrounding the approximate place was next scanned with a 2D laser profile measurement equipment, which gave the operator a detailed 3D representation of the area (Figure 2b), which included the bacterial colony.
A stainless steel needle was used to choose and place the colonies (Figure 2c). The selected colonies required to be put on MBT Biotarget 96 target plates before being examined using a MALDI Biotyper mass spectrometer (Figure 2d).
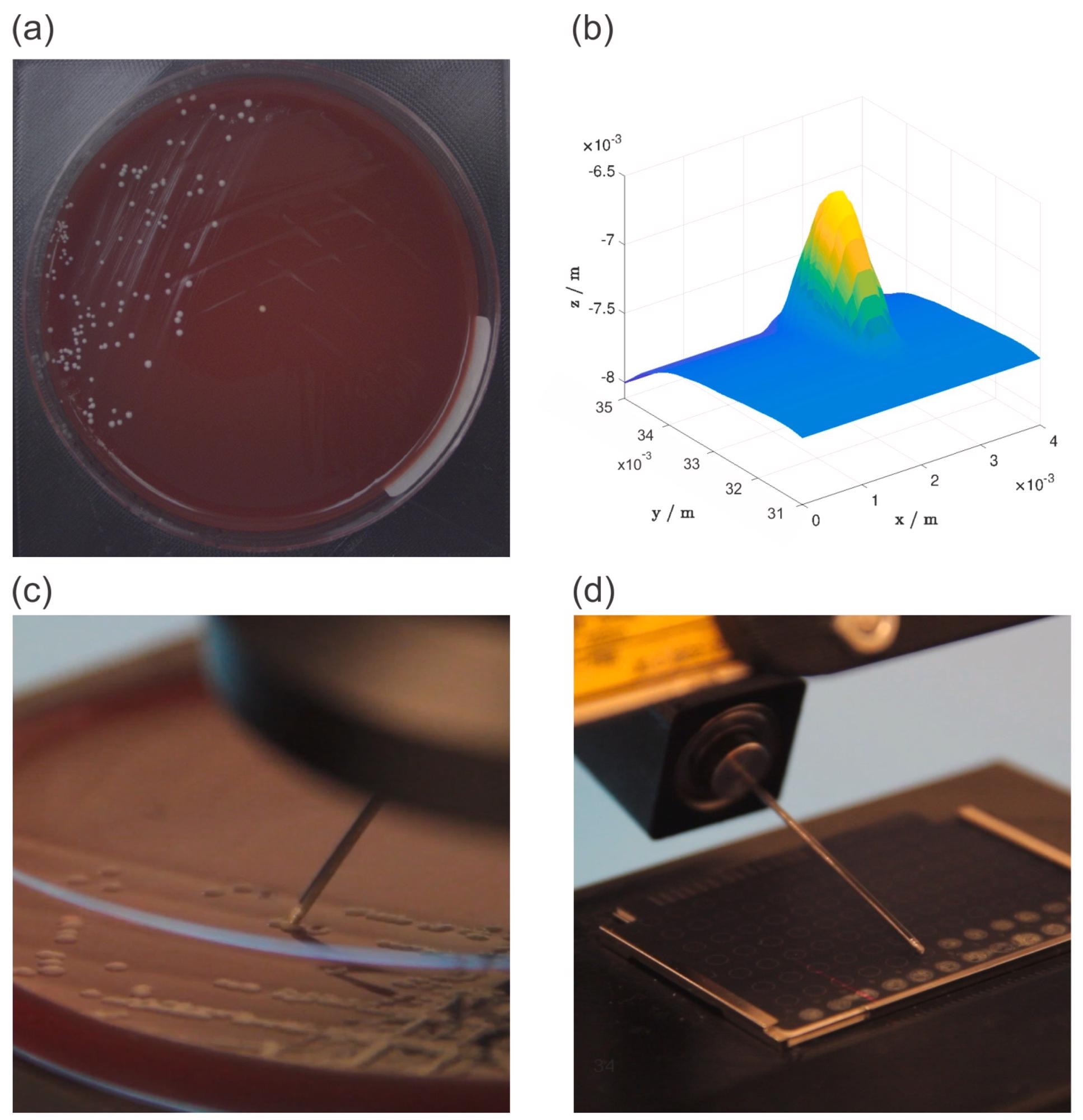
Figure 2. Overview of the experiment workflow: an image of a petri dish acquired with an RGB camera (a), a 3D model of the selected bacterial colony generated from 2D laser profiles (b), the bacterial colony picking process (c), and the bacterial colony deposition process (d). Image Credit: Baumkircher, et al., 2022
MATLAB Simulink 2019b was used to handle high-level control. The Panda robot, Nano17 force sensor, RGB camera, and 2D laser profile measurement system were used to link individual units, manage the experiment workflow, and preserve the measured data.
Learning from demonstration is a technique in which the operator performs the task rather than programming it. The so-called movement primitives are accustomed to observing demonstrations and learning from them. DMPs (dynamic movement primitives) are one of the various ways available.
However, for research purposes, encoding merely the movement position (as shown in Figure 3) was adequate.
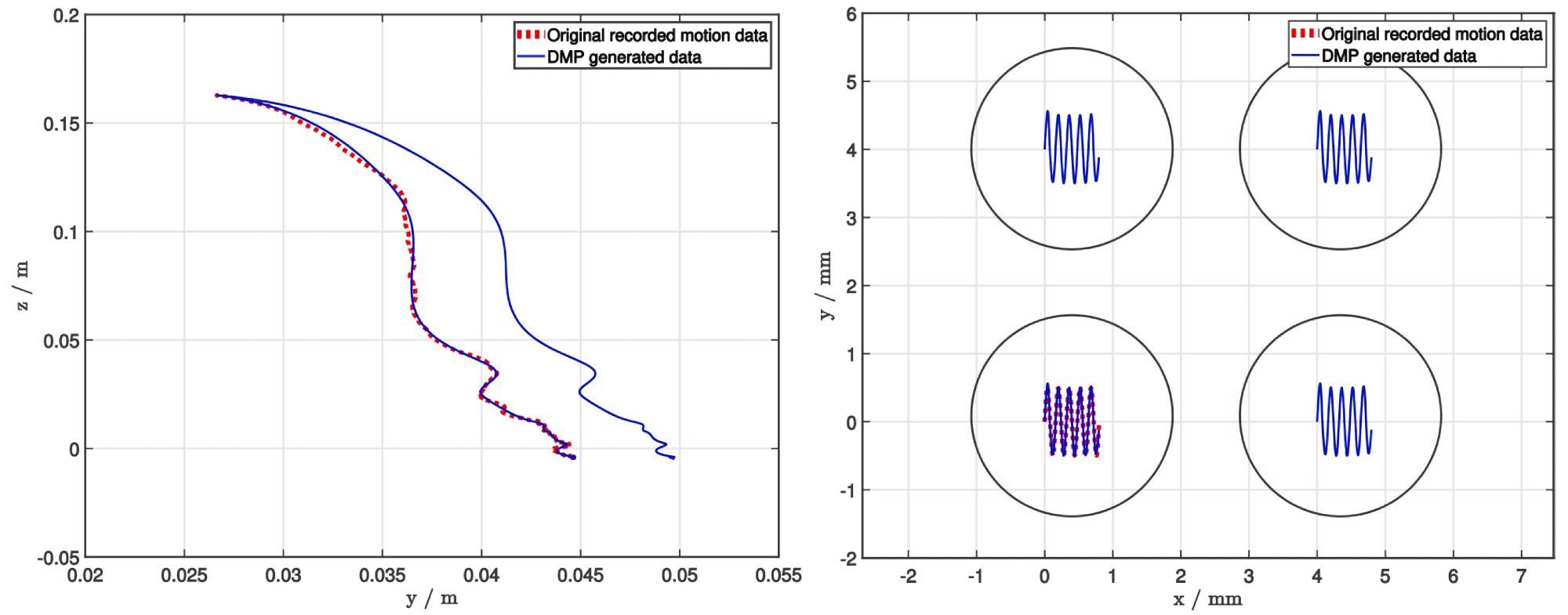
Figure 3. Using DMP to generalize demonstrated trajectories of the picking (left) and deposition (right) task. To demonstrate the quality of generalization, the end position of the picking process is modified, whereas both the start and end positions are modified for the deposition process. Image Credit: Baumkircher, et al., 2022
The decision to utilize a DMP was based on two factors. Initially, only one demonstration is required. This is crucial for collaborative robots in the laboratory since tasks must be taught in the shortest amount of time possible.
Second, the learned job can be adapted to varied start and finish places while maintaining a smooth trajectory. This is how researchers change the end position during the picking process, as well as the start and end positions during the deposition process, in the case study (Figure 3).
Researchers chose 56 colonies of two different species of bacteria cultivated on blood agar for the study. The operator presented the picking procedure at the start of the experiment, whereas the deposition process was developed programmatically. After that, a DMP was used to generalize both demonstrations (Figure 3).
Results and Discussion
Researchers obtained numerous colony 3D models before and after the picking process to assess the picking process. Researchers used a top-down heatmap to plot these models (Figure 4).
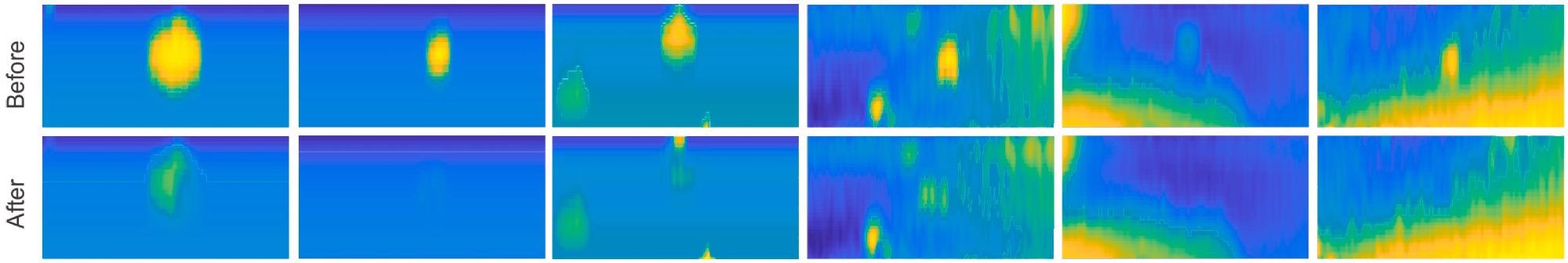
Figure 4. Overview of the picking process. Multiple different colonies are plotted before (top) and after (bottom) the picking process. Image Credit: Baumkircher, et al., 2022
They were categorized from left to right in Figure 4 and Table 1 depending on the original volume.
Table 1. Estimated colony volume change before and after the picking process. Source: Baumkircher, et al., 2022
| ID |
1 |
2 |
3 |
4 |
5 |
6 |
| Initial volume [mm3] |
1.44 |
0.78 |
0.46 |
0.11 |
0.06 |
0.02 |
| Final volume [mm3] |
0.41 |
-0.04 |
0.04 |
0.05 |
0.007 |
-0.02 |
| Relative change [%] |
-72 |
-105 |
-91 |
-54 |
-99 |
-193 |
The contact force is represented with the z location on the left plot of Figure 5. Since the data was collected from all 56 depositions, both are given with a variance plot.
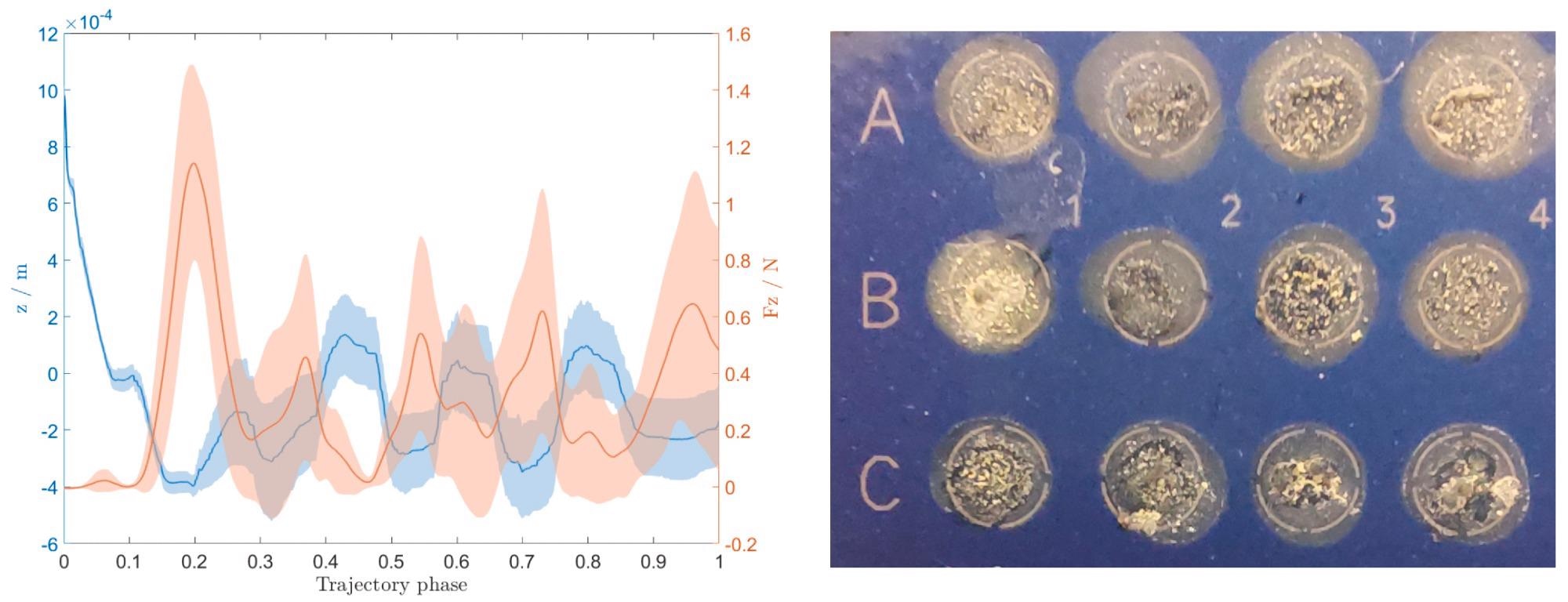
Figure 5. The contact force between the end–effector needle and MALDI target plate and the corresponding compensated z position (left), and an image of deposited colonies on the MALDI target plate after being covered with the matrix solution (right). Image Credit: Baumkircher, et al., 2022
Table 2 shows the average scores along with the related standard deviation. Since the low scores of incorrect samples affected the average score and, thus, the perception of overall performance, the scores were provided separately for accurately identified samples and all samples.
Table 2. Identification process results. Source: Baumkircher, et al., 2022
| |
Acinetobacter baumannii |
Staphylococcus epidermidis |
| No. of samples |
36 |
20 |
| No. of “score below 1.70” |
3 |
1 |
| No. of “no peaks found” |
2 |
0 |
| No. of total errors |
5 |
1 |
| Total errors [%] |
13.9 |
5 |
| Average identification score w/o errors |
2.05 ± 0.21 |
2.06 ± 0.13 |
| Average identification score with errors |
1.94 ± 0.54 |
2.04 ± 0.16 |
In six separate colonies, the selection process was studied. Researchers used the created 3D models of bacterial colony areas before and after the plucking to analyze them. It can be seen that in every case, the colony vanishes almost completely (Figure 4).
Furthermore, in most cases, the region where the colony once stood is not darker than its immediate surroundings. The picking was done correctly, with no needless removal of the agar layer beneath the colony. The volumes that have been measured further support all of this (Table 1).
The plot (Figure 5, left) shows that the force compensation works well during the deposition process.
However, if researchers look at the image of the deposited colonies (Figure 5, right), one can see that the samples are all deposited uniquely, even if they are all inside their allotted places. The majority are evenly scattered across the entire area, while a few are concentrated in smaller locations. Concentrated locations are undesirable since they can lead to less accurate identification.
Furthermore, more deposition sample treatments could enhance the identification outcomes. Researchers obtained the identification results shown in Table 2 when utilizing the direct spotting method (no extra formic acid). Nearly 11% of the samples could not be recognized.
Conclusion
Researchers employed a collaborative robot to see if kinesthetic training could be used for tasks that needed sub-millimeter accuracy. For the case study, researchers picked a bacterial colony selection and identification process employing mass spectrometry in a medical microbiology laboratory.
Researchers further created a specific robot end-effector that, in addition to picking and depositing, could also locate bacterial colonies, create a 3D model of them, and adjust contact forces.
An operator showed the tasks, which were then generalized using a DMP. Even though the colony detection, picking, and depositing workflow worked effectively, the total identification error rate was about 11%.
By utilizing kinesthetic teaching and DMPs, the team were successful in demonstrating fine tasks in a microbiological laboratory environment and generalizing them. Adjusting the deposited sample treatment, on the other hand, may allow for further system enhancements.
Journal Reference:
Baumkircher, A., Seme, K., Munih, M., Mihelj, M. (2022) Collaborative Robot Precision Task in Medical Microbiology Laboratory. Sensors, 22(8), p. 2862. Available Online: https://www.mdpi.com/1424-8220/22/8/2862/htm.
References and Further Reading
- Leber, A L (2020) Clinical Microbiology Procedures Handbook. In: John Wiley & Sons: Hoboken, NJ, USA.
- Buchan, B W & Ledeboer, N A (2014) Emerging technologies for the clinical microbiology laboratory. Clinical Microbiology Reviews, 27(4), pp. 783–822. doi.org/10.1128/CMR.00003-14.
- Patel, R (2013) Matrix-assisted laser desorption ionization–time of flight mass spectrometry in clinical microbiology. Clinical Infectious Diseases, 57(4), pp. 564–572. doi.org/10.1093/cid/cit247.
- Dadoun, R (2002) Case study: Automation’s impact on productivity and turnaround time. MLO Medical Laboratory Observer, 34(5), pp. 36–38.
- Bizzini, A., et al. (2010) Performance of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of bacterial strains routinely isolated in a clinical microbiology laboratory. Journal of Clinical Microbiology, 48(5), pp. 1549–1554. doi.org/10.1128/JCM.01794-09.
- Croxatto, A., et al. (2016) Laboratory automation in clinical bacteriology: What system to choose? Clinical Microbiology and Infection, 22(3), pp. 217–235. doi.org/10.1016/j.cmi.2015.09.030.
- Dauwalder, O., et al. (2016) Does bacteriology laboratory automation reduce time to results and increase quality management? Clinical Microbiology and Infection, 22(3), pp. 236–243. doi.org/10.1016/j.cmi.2015.10.037.
- Chudejova, K., et al. (2017) Validation of a novel automatic deposition of bacteria and yeasts on MALDI target for MALDI-TOF MS-based identification using MALDI Colonyst robot. PLoS ONE, 12(3), p. e0190038. doi.org/10.1371/journal.pone.0190038.
- Jones, P., et al. (1992) Integration of image analysis and robotics into a fully automated colony picking and plate handling system. Nucleic Acids Research, 20(17), pp. 4599–4606. doi.org/10.1093/nar/20.17.4599.
- Briner, D. R., et al. (2009) A Multi-Pin End-Effector for a Robotic Colony Picker. In: 2009 ASME Early Career Technical Conference, Tuscaloosa, AL, USA, 2–3 October 2009; pp. 222–228. Available at: https://www.uab.edu/home/
- Huang, C., et al. (2018) A colony picking robot with multi-pin synchronous manipulator. In: 2018 IEEE International Conference on Information and Automation (ICIA), Fujian, China, 11–13 August; pp. 7–12. doi.org/10.1109/ICInfA.2018.8812499.
- Fabritius, A., et al. (2018) Imaging-based screening platform assists protein engineering. Cell Chemical Biology, 25(12), pp. 1554–1561. doi.org/10.1016/j.chembiol.2018.08.008.
- Ravichandar, H., et al. (2020) Recent advances in robot learning from demonstration. Annual Review of Control, Robotics, and Autonomous Systems, 3, pp. 297–330. doi.org/10.1146/annurev-control-100819-063206.
- Argall, B. D., et al. (2009) A survey of robot learning from demonstration. Robotics and Autonomous Systems, 57(5), pp. 469–483. doi.org/10.1016/j.robot.2008.10.024.
- Schou, C., et al. (2013) Human-robot interface for instructing industrial tasks using kinesthetic teaching. In: IEEE ISR 2013, Seoul, Korea, 24–26 October; pp. 1–6. doi.org/10.1109/ISR.2013.6695599.
- Paraschos, A., et al. (2018) Using probabilistic movement primitives in robotics. Auton Robot, 42, pp. 529–551. doi.org/10.1007/s10514-017-9648-7.
- Steinmetz, F., et al. (2015) Simultaneous kinesthetic teaching of positional and force requirements for sequential in-contact tasks. In: 2015 IEEE-RAS 15th International Conference on Humanoid Robots (Humanoids), Seoul, Korea, 3–5 November 2015; pp. 202–209. doi.org/10.1109/HUMANOIDS.2015.7363552.
- Berio, D., et al. (2016) Learning dynamic graffiti strokes with a compliant robot. In: 2016 IEEE/RSJ International Conference on Intelligent Robots and Systems (IROS), Daejeon, Korea, 9–14 October 2016; pp. 3981–3986. doi.org/10.1109/IROS.2016.7759586.
- Kronander, K & Billard, A (2013) Learning compliant manipulation through kinesthetic and tactile human-robot interaction. IEEE Transactions on Haptics, 7(3), pp. 367–380. doi.org/10.1109/TOH.2013.54.
- Calinon, S., et al. (2014) Human—Robot skills transfer interfaces for a flexible surgical robot. Computer Methods and Programs in Biomedicine, 116(2), pp. 81–96. doi.org/10.1016/j.cmpb.2013.12.015.
- Huang, B., et al. (2017) A multirobot cooperation framework for sewing personalized stent grafts. IEEE Transactions on Industrial Informatics, 14(4), pp. 1776–1785. doi.org/10.1109/TII.2017.2773479.
- Liang, J., et al. (2017) Using dVRK teleoperation to facilitate deep learning of automation tasks for an industrial robot. In: 2017 13th IEEE Conference on Automation Science and Engineering (CASE), Xian, China, 20–23 August 2017; pp. 1–8. doi.org/10.1109/COASE.2017.8256067.
- Abbott, J. J., et al. (2003) Steady-hand teleoperation with virtual fixtures. In: 12th IEEE International Workshop on Robot and Human Interactive Communication, ROMAN 2003, Millbrae, CA, USA, 31 October–2 November 2003; pp. 145–151. doi.org/10.1109/ROMAN.2003.1251824.
- Bettini, A., et al. (2004) Vision-assisted control for manipulation using virtual fixtures. IEEE Transactions on Robotics, 20(6), pp. 953–966. doi.org/10.1109/TRO.2004.829483.
- Baumkircher, A., et al. (2021) Performance analysis of learning from demonstration approaches during a fine movement generation. IEEE Transactions on Human-Machine Systems, 51(6), pp. 653–662. doi.org/10.1109/THMS.2021.3107523.
- Ijspeert, A. J., et al. (2013) Dynamical movement primitives: Learning attractor models for motor behaviors. Neural Computation, 25(2), pp. 328–373. doi.org/10.1162/NECO_a_00393.
- Calinon, S., et al. (2007) On learning, representing, and generalizing a task in a humanoid robot. IEEE Transactions on Systems, Man, and Cybernetics, Part B (Cybernetics), 37(2), pp. 286–298. doi.org/10.1109/TSMCB.2006.886952.
- Huang, Y., et al. (2019) Kernelized movement primitives. The International Journal of Robotics Research, 38(7), pp. 833–852. doi.org/10.1177%2F0278364919846363.
- Paraschos, A., et al. (2013) Probabilistic movement primitives. In: Advances in Neural Information Processing Systems; MIT Press: Cambridge, MA, USA.