Image Credit: University of Surrey.
The HCPL on the Human Protein Atlas was tested by the team, who discovered it to be the most precise tool for determining the location of proteins within individual cells.
To understand how proteins work inside cells, scientists need to study where they are located, but this can be a time-consuming and complicated process. HCPL makes this process easier.
Miroslaw Bober, Professor and leader of the HCPL Project, University of Surrey
Bober added, “This program uses a deep-learning model to quickly and accurately identify subcellular structures where proteins are present inside individual cells. We are hopeful that HCPL can help scientists study how proteins work and develop new treatments for diseases.”
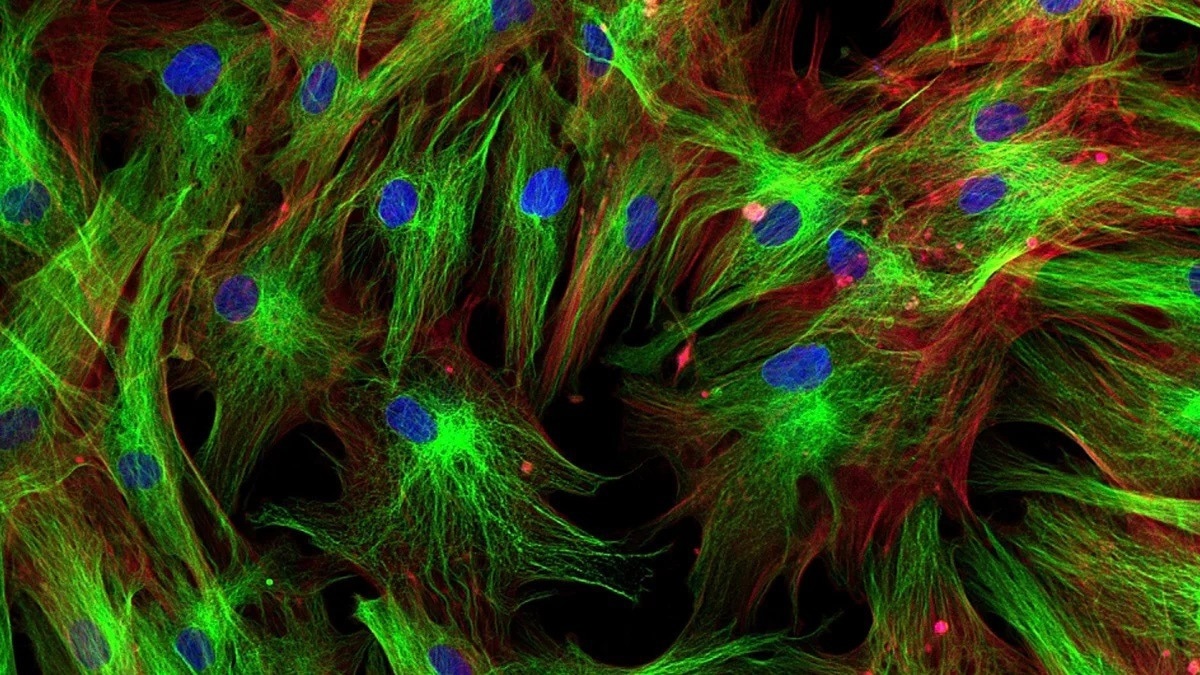
Spatial proteomics is known as a research area that studies the distribution of proteins in tissues or cells by making use of a combination of experimental methods and computational approaches. Fluorescence microscopy is a general method in this field where proteins have been tagged physically together with fluorescent markers.
AI helps map the proteins onto separate cell compartments (subcellular structures or organelles). This aids researchers in comprehending the roles and functions of proteins and possibly discloses the complicated inner workings of cells.
HCPL was developed in collaboration with ForecomAI, a research and development company having world-class expertise in machine and deep learning offering solutions in biosciences and healthcare.
Proteins play a key role in most cellular processes crucial to our survival. Unraveling protein distributions and interactions within individual cells is vital to understanding their functions and indispensable to developing new treatments.
Dr. Amaia Irizar, Director, ForecomAI
Irizar added, “Our work with the University of Surrey enables scaling up of this process and opens new frontiers. The partnership between Surrey and ForecomAI has been a successful interdisciplinary collaboration in scientific research, paving the way for further initiatives.”
Journal Reference:
Husain, S. S., et al. (2023) Single-cell subcellular protein localization using novel ensembles of diverse deep architectures. Communications Biology. https://doi.org/10.1038/s42003-023-04840-z.